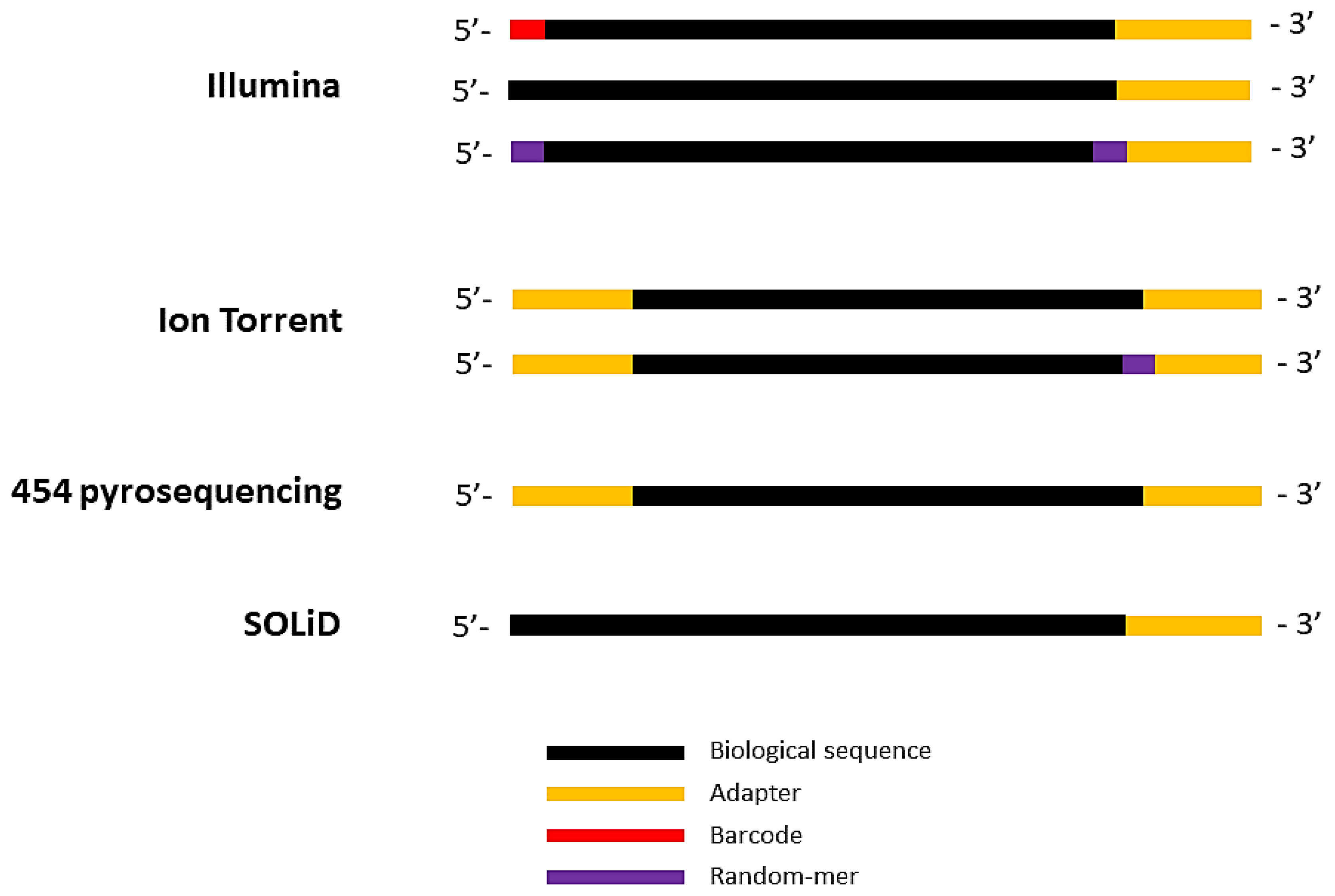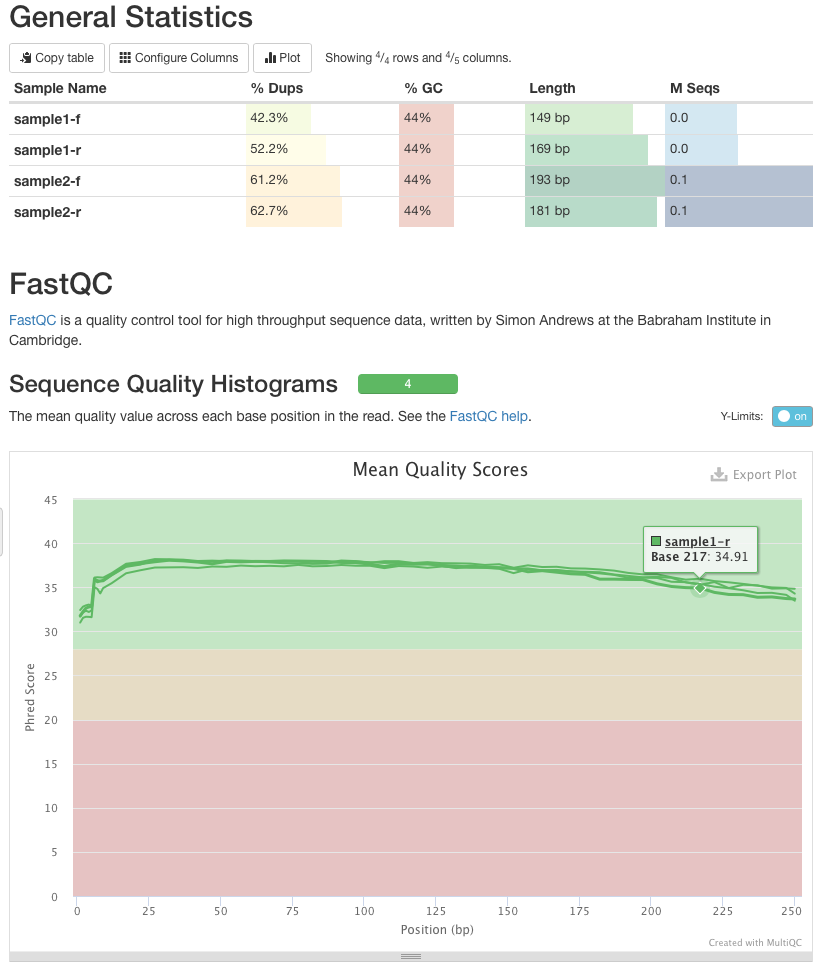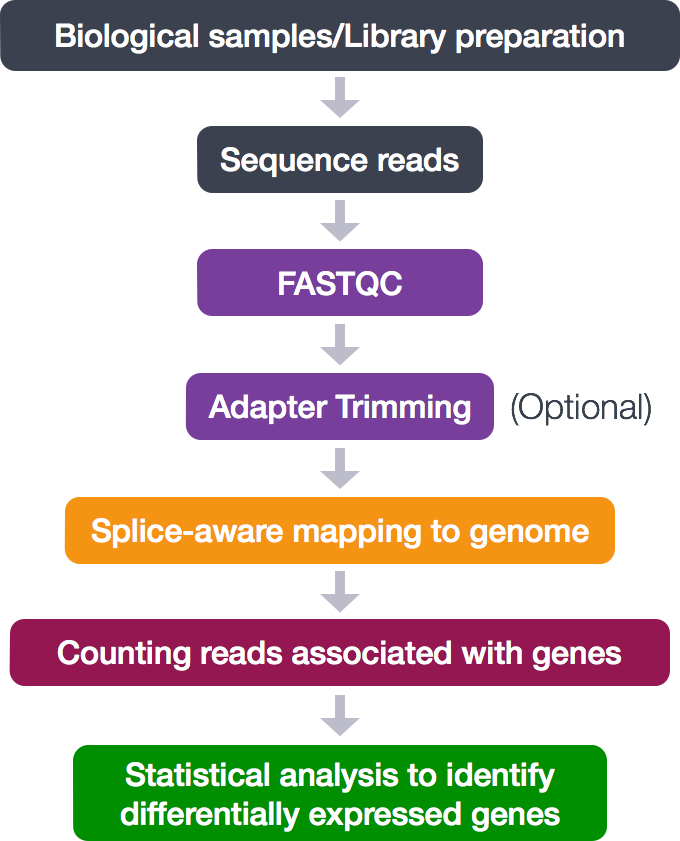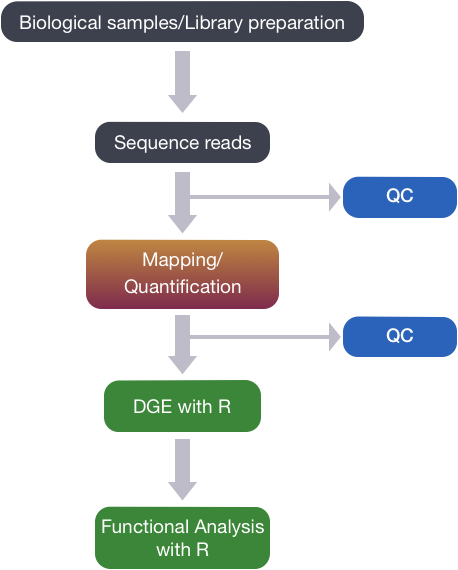
Quality control: Assessing FASTQC results | Introduction to RNA-Seq using high-performance computing - ARCHIVED
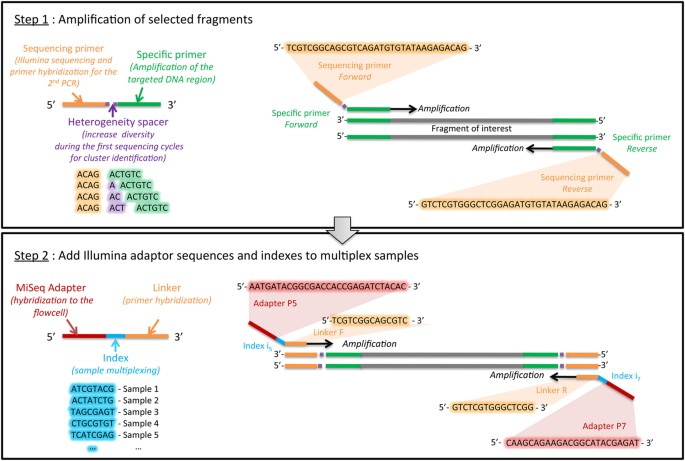
High-throughput sequencing of multiple amplicons for barcoding and integrative taxonomy | Scientific Reports
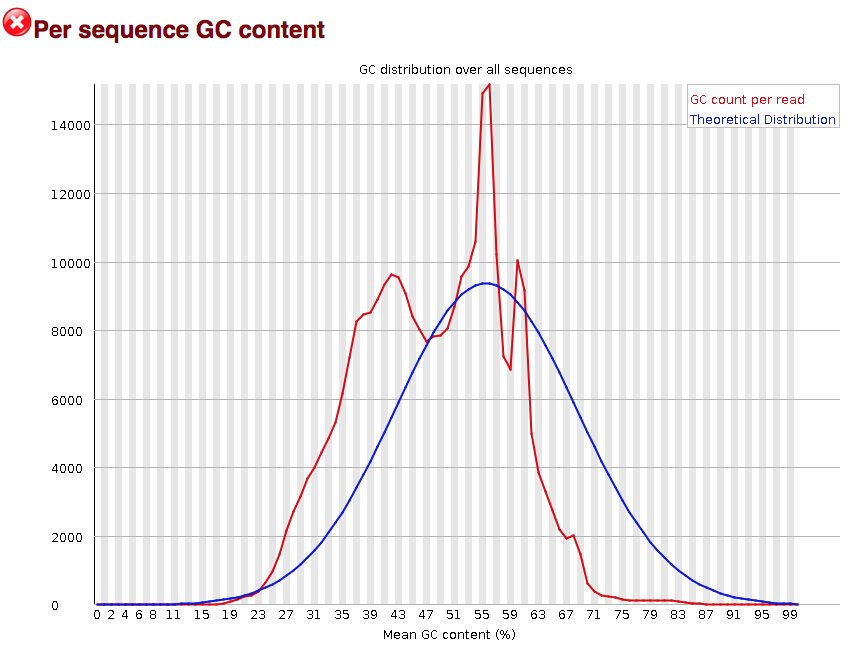
Quality control: Assessing FASTQC results | Introduction to RNA-Seq using high-performance computing - ARCHIVED
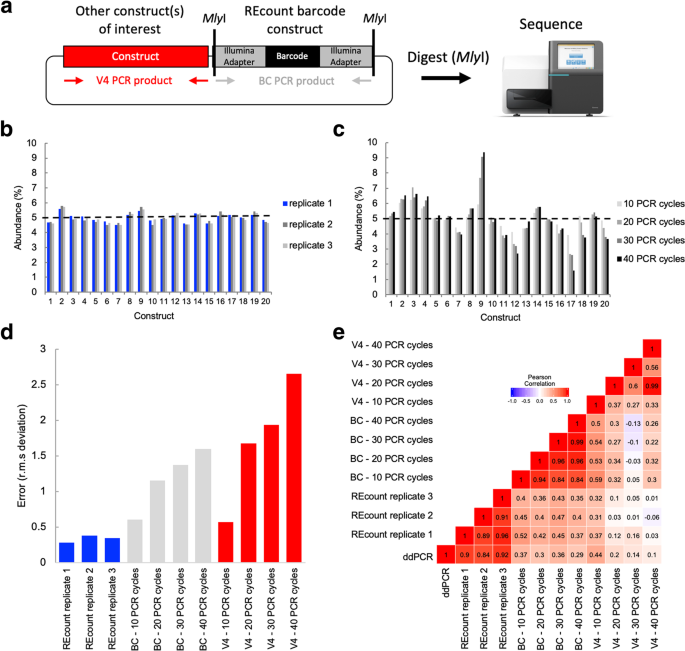
Measuring sequencer size bias using REcount: a novel method for highly accurate Illumina sequencing-based quantification | Genome Biology | Full Text