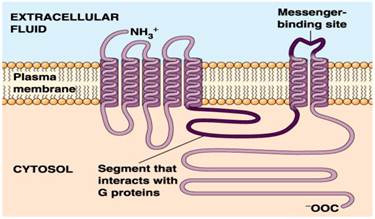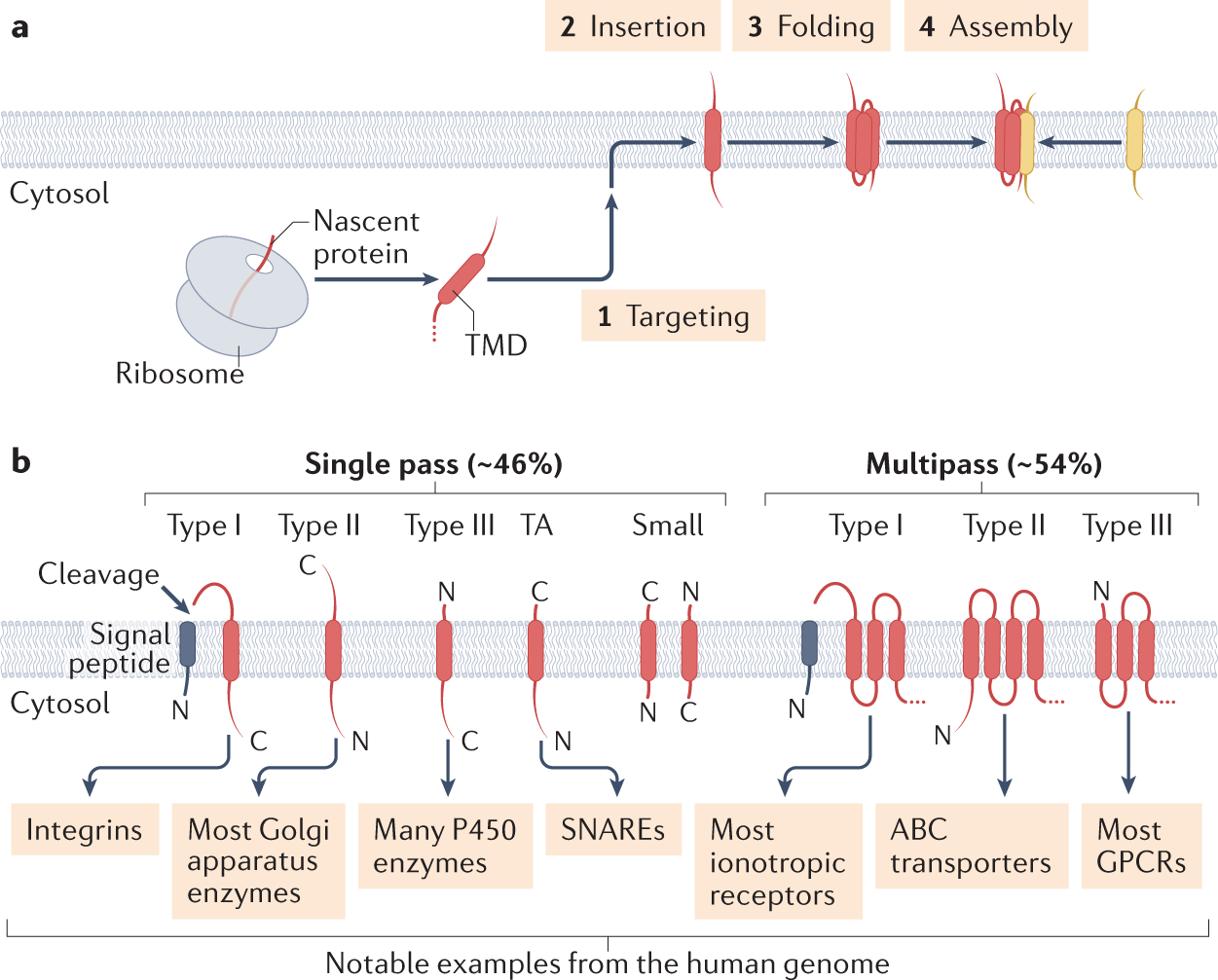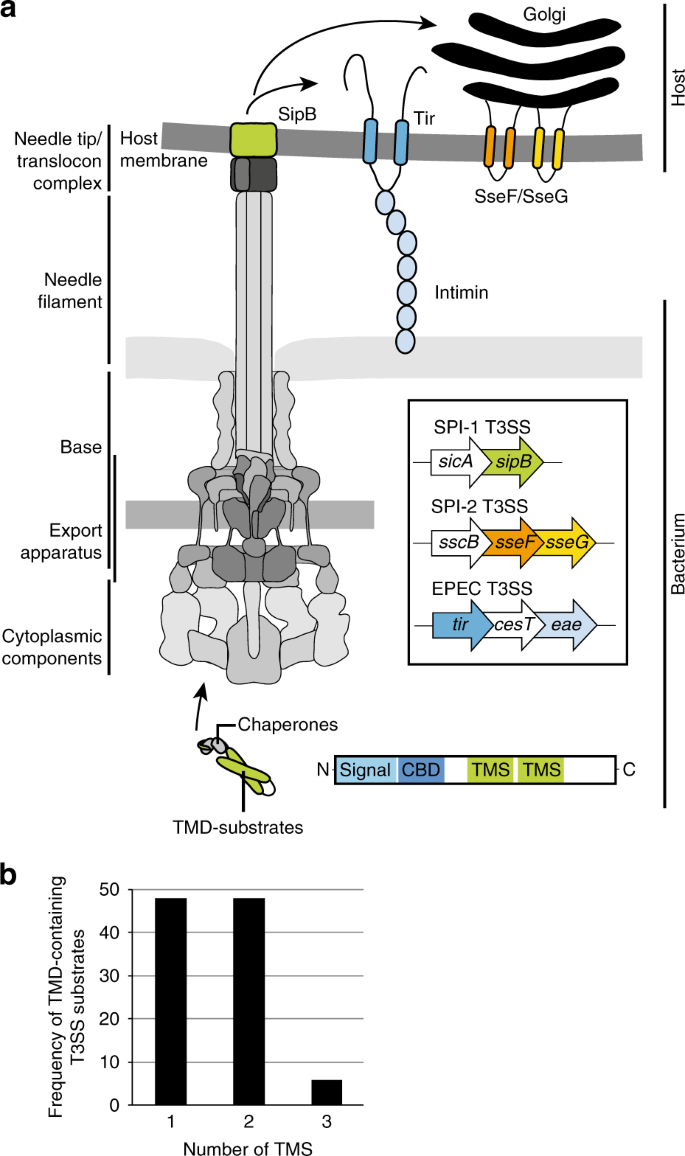
Revealing the mechanisms of membrane protein export by virulence-associated bacterial secretion systems | Nature Communications

Getting to know each other: PPIMem, a novel approach for predicting transmembrane protein-protein complexes - ScienceDirect

Interplay between hydrophobicity and the positive-inside rule in determining membrane-protein topology | PNAS

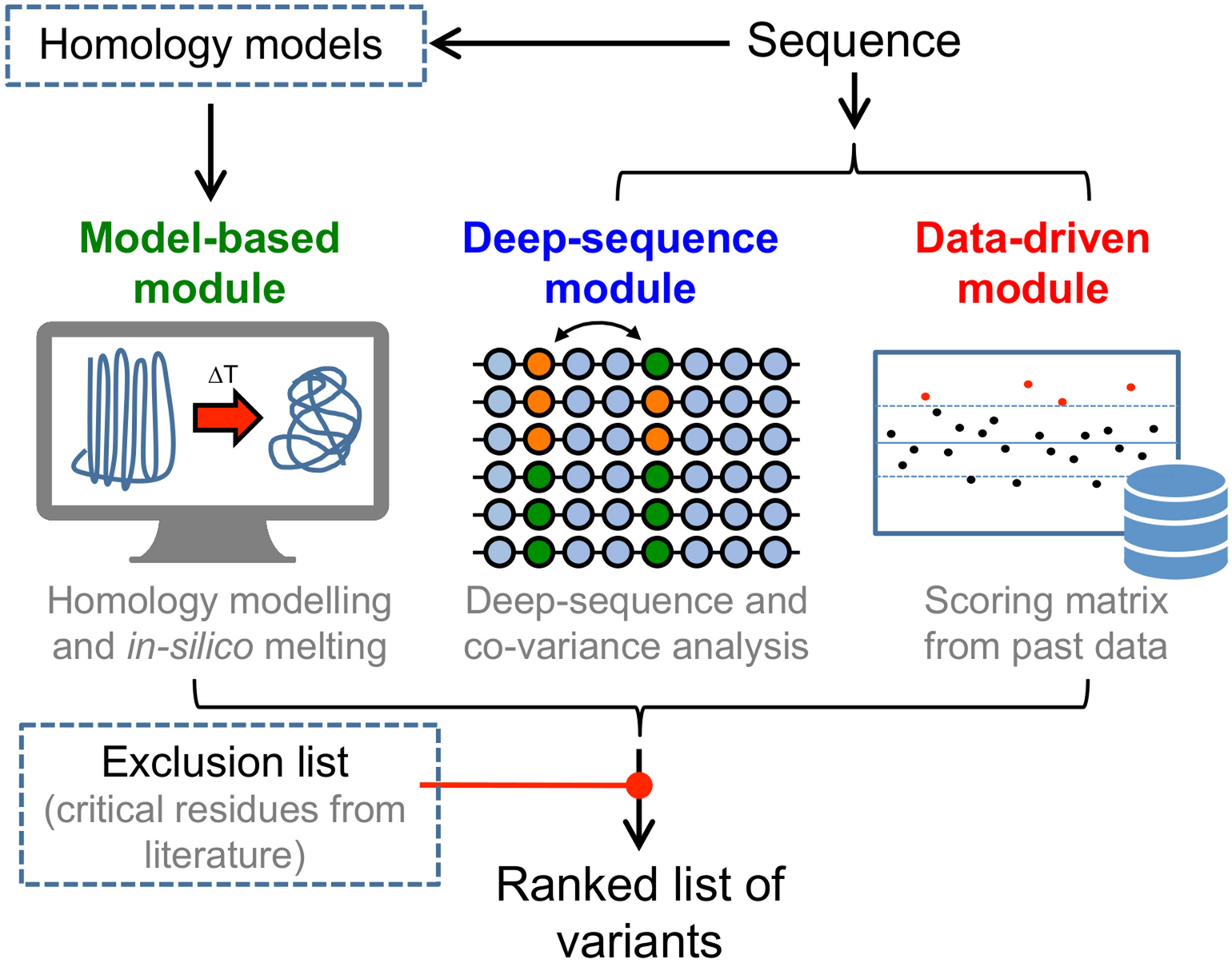
MutTMPredictor: Robust and accurate cascade XGBoost classifier for prediction of mutations in transmembrane proteins - ScienceDirect

Topology Prediction Improvement of α-helical Transmembrane Proteins Through Helix-tail Modeling and Multiscale Deep Learning Fusion - ScienceDirect

Methods for Systematic Identification of Membrane Proteins for Specific Capture of Cancer-Derived Extracellular Vesicles - ScienceDirect
Membrane protein contact and structure prediction using co-evolution in conjunction with machine learning | PLOS ONE
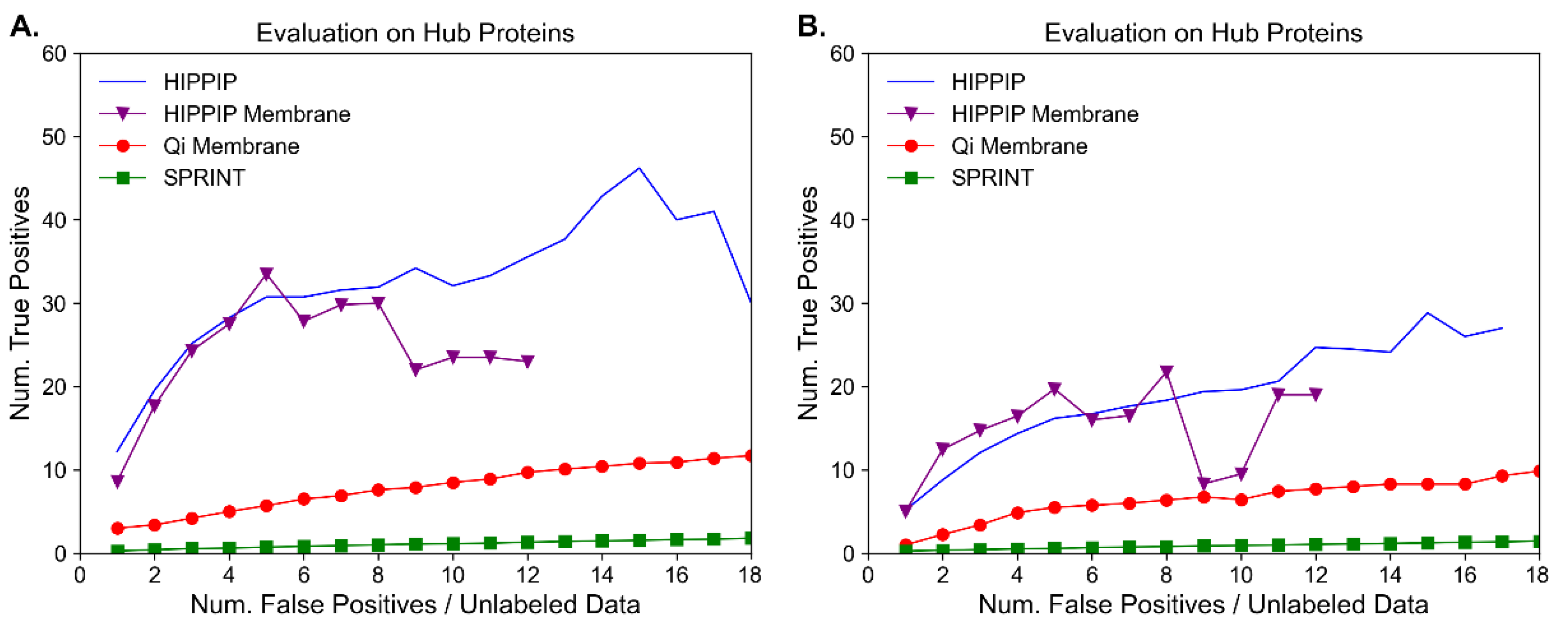
Molecules | Free Full-Text | Benchmark Evaluation of Protein–Protein Interaction Prediction Algorithms | HTML
Membrane protein contact and structure prediction using co-evolution in conjunction with machine learning | PLOS ONE

Interplay between hydrophobicity and the positive-inside rule in determining membrane-protein topology | PNAS
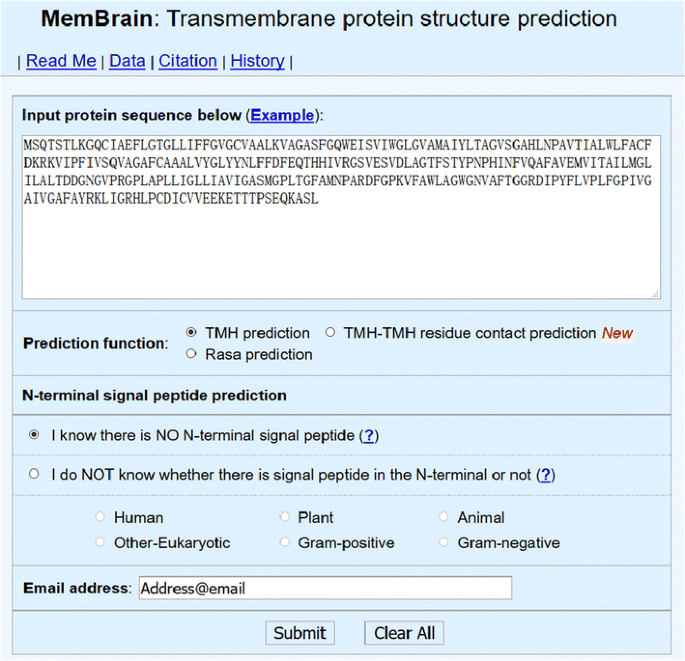
MemBrain: An Easy-to-Use Online Webserver for Transmembrane Protein Structure Prediction | SpringerLink
A lipophilicity-based energy function for membrane-protein modelling and design | PLOS Computational Biology

Interplay between hydrophobicity and the positive-inside rule in determining membrane-protein topology | PNAS
Membrane protein contact and structure prediction using co-evolution in conjunction with machine learning | PLOS ONE
MemBrain: An Easy-to-Use Online Webserver for Transmembrane Protein Structure Prediction | SpringerLink

MemBrain: An Easy-to-Use Online Webserver for Transmembrane Protein Structure Prediction | SpringerLink
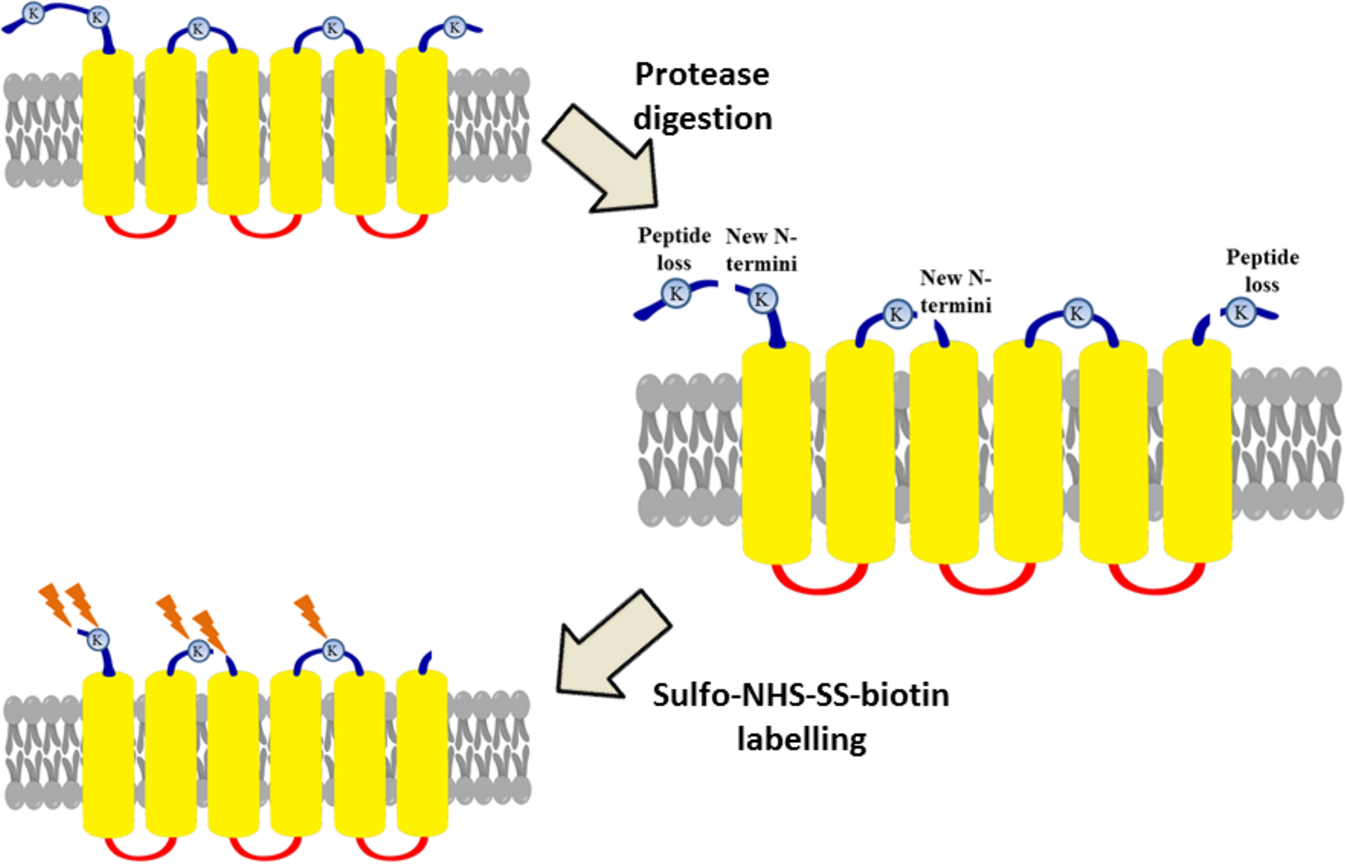
Partial proteolysis improves the identification of the extracellular segments of transmembrane proteins by surface biotinylation | Scientific Reports